[ad_1]
reagent
Antibodies against the following proteins were purchased from Proteintech. Snail1 (13099-1-AP, 1:1000 dilution). RhoA (10749-1-AP, 1:1000 dilution); α-SMA (14395-1-AP, 1:1000 dilution); Col1a1 (14695-1-AP, 1:1000 dilution); Flag (66008-4- Ig, 1:1000 dilution). HA (51064-2-AP, 1:1000 dilution); Ubiquitin (10201-2-AP, 1:1000 dilution); GAPDH (60004-1-Ig, 1:1000 dilution); β-actin (81115-1- RR, 1:1000 dilution). GTP-RhoA antibody was purchased from NewEast Biosciences (26904, 1:1000 dilution). FBXL8 antibody was purchased from Santa Cruz (sc-390582, 1:1000 dilution). FBXL8 siRNA was purchased from Ribobio (Guangzhou, China). Adenoviruses overexpressing FBXL8 and green fluorescent protein (GFP) were purchased from Vigenebio (Rockville, MD, USA). CCG-1423 (HY-13991), bafilomycin A1 (HY-100558), MG-132 (HY-13259), cycloheximide (CHX, HY-12320), and TGFβ (HY-P70543) were purchased from MCE .
Rat model of MI
Six-week-old male Sprague-Dawley rats (weighing 200–250 g) were purchased from JieSiJie Laboratory Animal Co., Ltd. (Shanghai, China). All experimental rats were kept in standard cages (12 h light/dark cycle; temperature, 21 ± 1 °C; humidity, 55–60 °C). Briefly, before grouping, each rat was assigned one consecutive random number from a random number table in order of body weight. He then ranged all the rats from small to large by random numbers and assigned every six of his consecutive rats to one group. Set the significance level (α) with 0.05 and power (1”)β) 80% and determine the sample size according to the preliminary experiment. No samples or animals were excluded from data analysis. The exact number of groups is stated in the figure legend. All experimental procedures were performed in accordance with institutional animal care. The Institutional Animal Care and Use Committee of Shanghai General Hospital approved the animal protocol in this study. The researchers were blinded to the different group assignments when assessing the results. As previously described, the left anterior descending (LAD) coronary artery was permanently ligated to establish the MI model. [19]. Briefly, 3% pentobarbital was used to anesthetize the rats, and the rats underwent endotracheal intubation and respiratory support. Subsequently, the rats’ hearts were exposed and the LAD was ligated with a 7-0 monofilament nylon suture, and the sham group underwent the same surgery except that the LAD was ligated.
echocardiography
Cardiac systolic function was assessed by two-dimensional and M-mode at 4 weeks after surgery using a Vevo 770 system (VisualSonics, Toronto, Canada). Left ventricular (LV) end-systolic internal dimensions (LVIDs) and end-diastolic LV internal dimensions (LVIDd) were measured using left ventricular parasternal long-axis views. Ejection fraction (EF%) and fractional shortening (FS%) were calculated using the following formulas: \({\rm{EF}} \% =[({{\rm{LVIDd}}}^{3}{\boldsymbol{-}}{{\rm{LVIDs}}}^{3})/{\rm{LVIDd}}^{3}]\times 100{;}{\rm{FS}} \% =[({\rm{LVIDd}}-{\rm{LVIDs}})/{\rm{LVIDd}}]\Hundredfold\).
histology
Rat hearts were perfused and fixed in 4% paraformaldehyde for 24 h. Hearts were then cut horizontally, embedded in paraffin, and sliced into 5 μm thick slices. Scar tissue area and infarct wall thickness in heart sections were determined by Masson’s trichrome staining and evaluated by Image J software.
Fluorescence immunohistochemistry
Heart sections were deparaffinized, rehydrated, and antigen retrieved. Fibroblasts or cardiomyocytes were fixed with 4% formaldehyde and permeabilized with 0.1% Triton X-100. Heart sections or cells were blocked with 10% bovine serum albumin (BSA) for 15 min and incubated with primary antibodies overnight at 4 °C. The following primary antibodies were used for immunofluorescence staining: FBXL8 (sc-390582, 1:100 dilution), Snail1 (13099-1-AP, 1:200 dilution), α-SMA (14395-1-AP, 1 :200 dilution), Col1a1 (14695-1-AP, 1:200 dilution). The next day, slides or cells were incubated with Alexa Fluor 488 or 555-conjugated secondary antibodies and DAPI for 15 min and 90 min at room temperature.
Cell culture and transfection
The human lung fibroblast (HLF) MRC-5 cell line was obtained from American Type Culture Collection (ATCC) (Manassas, VA, USA), and the human dermal fibroblast (HDF) CC-2511 cell line was obtained from Clonetics (Sun, CA, USA). State Diego). As previously described, he isolated and cultured neonatal rat CF from 1-2 day old Sprague-Dawley rats. [19]. CFs were starved with serum-free DMEM overnight and then stimulated with TGFβ (10 ng/ml) for 24 h. CFs were transfected with the indicated plasmids 36 hr before TGFβ stimulation.
Cell proliferation and migration assay
Cell proliferation capacity was assessed using the CCK-8 kit (MedChemExpress, HY-K0301) according to the manufacturer’s protocol. The absorbance of each well was measured at 450 nm using a microplate reader. Cell migration was measured by transwell assay. Briefly, CFs were seeded in the upper chamber of a 24-well plate in 100 μL of serum-free medium incubated with or without TGFβ, and DMEM medium containing 10% FBS was added to the lower chamber. The experimental group was transfected with FBXL8 plasmid or FBXL8 siRNA, and the control group was transfected with empty plasmid or control scramble. The next day, CFs were fixed with 4% formaldehyde, stained with hematoxylin, and photographed.
Cell contraction assay
A total of 100,000 CFs were seeded in a collagen matrix and transferred to a 24-well plate coated with 1% BSA. Serum-free DMEM with or without TGFβ was added to the culture wells with the collagen gel released from the edges. The gels were scanned on day 3 and the surface diameter of the gels was measured using ImageJ software.
Plasmid construction
Plasmid encoding full-length human Fbxl8 with HA tag HA-Fbxl8 aa 8-45 deletion mutant (8-45μF), HA-Fbxl8 aa 46-154 deletion mutant (46-154μC1), HA-Fbxl8 aa 155-254 deletion mutant (155-254ΔC2), HA-Fbxl8 aa 255-374 deletion mutant (255-374ΔC3), Flag-Snail1 and Flag-Snail1 fragment (C-terminal amino acids 152-264 and N-terminal Amino acids 1-151) were purchased from WZ biosciences.
polymerase chain reaction
Total RNA was extracted using Trizol reagent (Invitrogen) according to the product manual. cDNA was synthesized by PrimeScript® RT reagent Kit (TAKARA), and qPCR was performed using SYBR green PCR master mix (Roche) according to the product manual. FBXL8 mRNA levels in Figure 1C were normalized by mRNA for β-actin expression. Relative formula calculated by 2“”ΔΔCT. PCR products were run on an agarose gel and then separated on a 6% non-denaturing polyacrylamide gel. Quantification of band density was performed using ImageJ software. Snail1 mRNA in Figure 5A was normalized by GAPDH expression mRNA. Supplementary Table 1 lists the primer pairs used in this study.
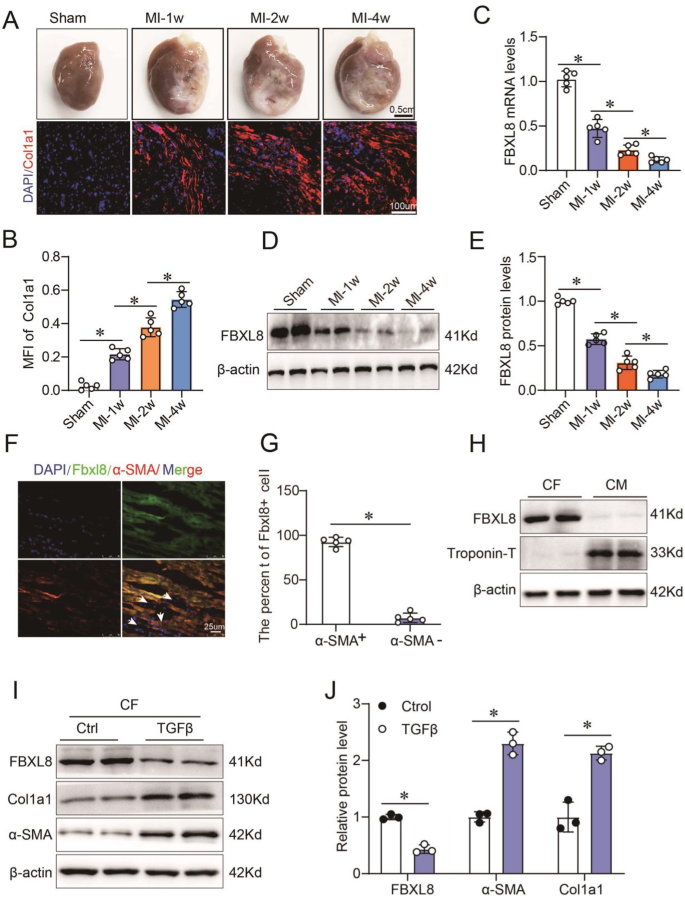
a, B Macroscopic specimens, immunofluorescence (a), quantitative (B) Col1a1 levels in the myocardium of rats undergoing sham or MI surgery at the indicated time points (n≧5). C–E mRNA (C) and proteins (D, E) FBXL8 expression in sham and MI hearts at 1 week (1 week), 2 weeks, and 4 weeks after surgery (n≧5). F, G Immunofluorescence images (F) and quantitative (G) Staining of LV sections from WT rats showing colocalization of FBXL8 expression (green) and α-SMA positive cells (red). Scale bar: 25μm. H Western blot of FBXL8 in cardiac fibroblasts (CF) and cardiomyocytes (CM). I, J Representative western blot (I) and quantitative (J) Cola1 protein levels in CFs treated with FBXL8, α-SMA, and TGFβ (10 ng/ml) for 48 h (n→3). Data are presented as mean ± SD. *P≦0.05.
western blot
Ventricular tissue and cells were lysed using RIPA buffer (Beyotime Biotechnology). Tissue or cell lysates were centrifuged at 12,000×, and the supernatant was extracted.g 10 minutes at 4°C. Protein extracts were separated using SDS-PAGE gels and the separated proteins were transferred to his PVDF membrane. PVDF membranes were blocked with 5% milk for 1 h at room temperature and then incubated with the indicated primary antibodies overnight at 4 °C. On the second day, the PVDF membrane was washed with TBST for 10 min and incubated with secondary antibody for 60 min at room temperature. Protein bands were captured using a Tanon 5200. β-actin or GAPDH was used to normalize specific protein levels.
Immunoprecipitation (IP) assay
All IP tests were performed using commercially available kits (#635696;Takara). Cultured HEK293T cells were transiently transfected with HA-FBXL8 and Flag-Snail1 plasmids and cultured for 48 h. Cells were then lysed in ice-cold IP buffer containing protease inhibitor cocktail and centrifuged at 13,000×.g 15 minutes. The resulting cell lysates were precleared with protein A/G agarose beads for 3 h and incubated with the indicated antibodies overnight at 4 °C. Immune complexes were collected after washing with cold IP buffer and subjected to immunoblotting using the indicated primary antibodies and corresponding secondary antibodies.
Ubiquitination assay
Cultured CFs were lysed in SDS lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% SDS) containing protease inhibitors (Beyotime, P1010). Lysates were denatured by heating at 95 °C for 5 min, diluted 10-fold with lysis buffer, and centrifuged at 20,000×.g 30 minutes at 4°C. Supernatants were immunoprecipitated with the indicated antibodies, followed by Western blotting.
statistical analysis
Data are presented as mean ± SD. SPSS version 16.0 was used to analyze the data. The Shapiro-Wilk test was used to assess the Gaussian distribution of the data.Two-tailed student without a pair t-test was used for comparisons between two groups. Multiple group comparisons were analyzed using one-way analysis of variance followed by post hoc Tukey tests. P≦0.05 is considered a statistically significant difference.
[ad_2]
Source link