[ad_1]
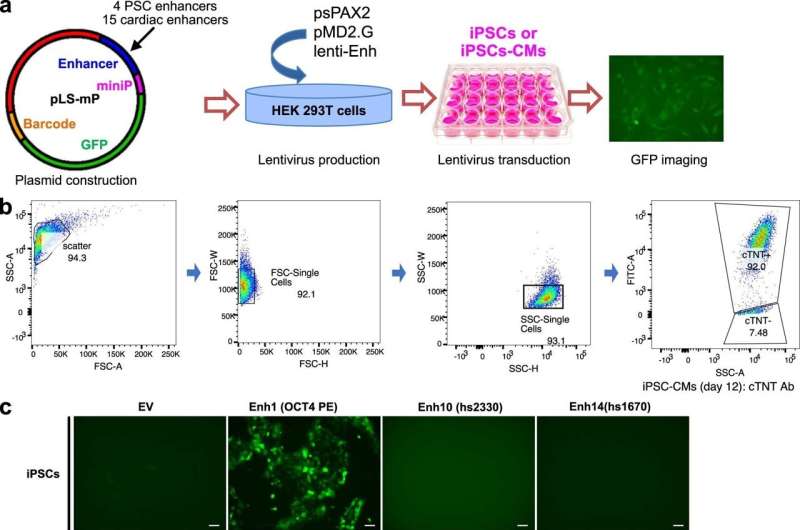
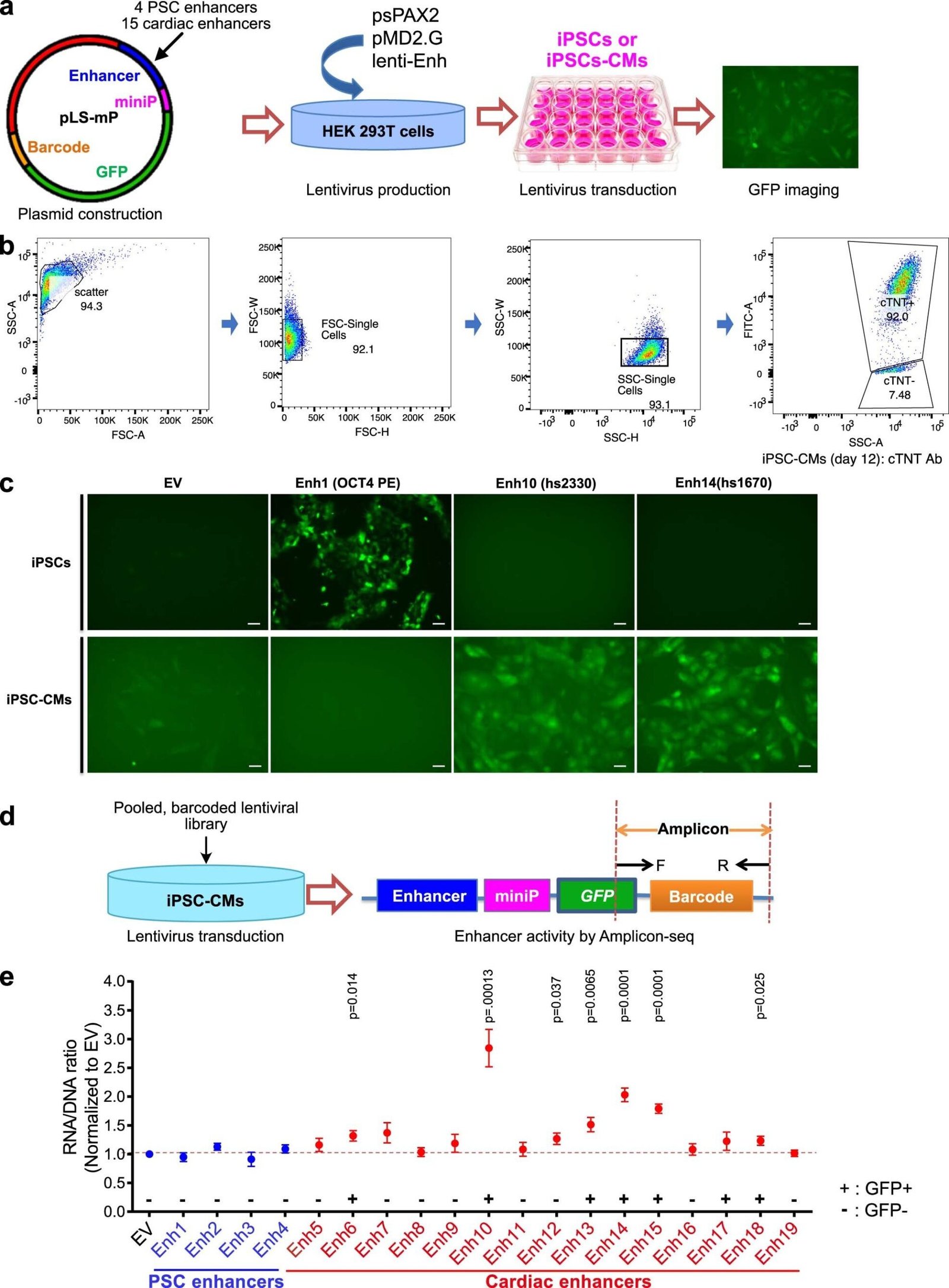
Establishment of the lentiMPRA platform to test cardiac enhancer activity in iPSC-CMs. a. Pilot experiment strategy to test the lentiviral reporter assay in iPSC-CM. b. Flow cytometry analysis of cTNT+ iPSC-CMs at day 12 of differentiation. c. Activation of PSC-specific and cardiac enhancers. d. Pilot experiment strategy for measuring enhancer activity by Amplicon-seq e. Enhancer activity of PSC enhancers (Enh1-4) and cardiac enhancers (Enh 5-19). credit: natural genetics (2024). DOI: 10.1038/s41588-024-01669-y
For decades, researchers have been slowly uncovering the genetic causes of congenital heart disease (CHD). Approximately 45% of CHD cases have an identifiable cause, such as chromosomal abnormalities, genetic mutations that affect protein-coding genes, or environmental factors. What about the rest of CHD?
Although non-coding DNA elements have long been thought to be involved in CHD, few variants have been identified as causative. A new study from the laboratory of cardiologist William Pugh, MD, at Boston Children’s Hospital provides the strongest evidence yet that non-coding DNA contributes to CHD. It also establishes a pipeline for finding and validating variants.
“It’s very difficult to determine which non-coding variants are actually involved in heart disease and which are just innocent bystanders,” said Dr. Chairman Poo says: “The average person has about 70 new variants (not shared with their parents) in non-coding regions. And when you compare people with and without heart disease, they have the same number of variants. I am.”
As explained in the article published in natural genetics, Pu and colleagues used data from the multicenter Pediatric Cardiac Genomics Consortium (PCGC), led by Jane Neuberger, MD, MPH, at Boston Children’s Hospital. Amy Roberts, MD. and Wendy Chan, MD.
The consortium enrolls more than 13,000 CHD patients (about one-fifth of them at Boston Children’s Hospital) and more than 18,000 family members. More than 3,000 patients have had their complete genome sequenced, and Pu’s lab has obtained whole-genome sequence data for 750 of her patients and their parents.
Narrow down the causative variant
To eliminate non-coding mutations that were likely to be background “noise” rather than the cause, the researchers applied statistical filters. They also focused on novel variants that are likely to be the cause.
This left nearly 7,000 mutations in non-coding DNA regions in CHD patients. To investigate the function of the mutants, the team created a high-throughput system to test the effects of the mutants on the expression of neighboring genes.
“Of the 7,000 mutants we tested, we found 403 that affected the activity of transcriptional enhancers,” Pu said. “These would be good candidates for variants that may actually contribute to congenital heart disease.”
They then painstakingly introduced the 10 non-coding variants into the appropriate locations within the genome of normal human stem cells. When the stem cells were instructed to form cardiomyocytes, four of the 10 mutants changed the expression of adjacent genes.
“These genes were already known to cause congenital heart disease when mutated,” Pugh said. “In mutants, the enhancer is no longer able to perform its original function, resulting in low gene expression levels, or it turns a region that should not be an enhancer into an enhancer, activating the gene at the wrong time or in the wrong place. Both scenarios can lead to heart disease.”
As a second test, we performed single-cell sequencing of cardiomyocytes derived from stem cells carrying the four mutations. These cells also have altered gene expression profiles, consistent with their potential to cause CHD.
Future directions of CHD genetics
Pu believes that hundreds of additional non-coding mutations may be involved in structural heart disease and help explain many cases of CHD that are negative on exome sequencing. Co-lead author Sarah Morton, MD, of the Department of Neonatal Medicine, specializes in researching the genetic causes of CHD. Using data from her study, she created an algorithm to prioritize the variants most likely to be associated with her CHD for further investigation.
“When evaluating coding gene variants, such as missense variants that change a single amino acid, we want to know how likely they are to alter protein expression,” Morton says. “There are relatively few tools to predict the functional impact of non-coding mutations, especially those that may be associated with pediatric disease.”
The researchers hope to expand their search beyond cardiomyocytes to other cell types, such as neural crest cells and endocardial cells.
In the future, the results of this study could be used to stratify patient risk, predict surgical outcomes, and even more distantly, change the trajectory of structural heart disease during pregnancy and after birth. There is sex.
“It’s completely unexplored territory,” Pugh said. “What we discovered in this study is just the tip of the iceberg.”
For more information:
Feng Xiao et al. Functional dissection of human cardiac enhancers and non-coding novel variants in congenital heart disease, natural genetics (2024). DOI: 10.1038/s41588-024-01669-y
Provided by Boston Children’s Hospital
Quote: In genetics of congenital heart disease, non-coding DNA fills some gaps (April 8, 2024), https://medicalxpress.com/news/2024-04-genetics-congenital-heart-disease Retrieved April 8, 2024 from -noncoding. html
This document is subject to copyright. No part may be reproduced without written permission, except in fair dealing for personal study or research purposes. Content is provided for informational purposes only.
[ad_2]
Source link